Cancer treatment has significantly evolved since the approval of the first companion diagnostic, HercepTest, in 1998. Both targeted therapies and immunotherapies have revolutionized cancer treatment, benefiting greatly from precision biomarker strategies which enhance outcomes in selected patient populations. However, we are beginning to reach the limits of existing biomarker-driven treatment efficacy and development.
Broadly, tissue-based biomarkers fall into two main categories, each with innate limitations which present challenges for drug development. Molecular biomarkers (e.g. EGFR, MSI, etc.) indicate a specific genetic alteration that leads to an abnormal phenotype but they fail to consider the heterogeneity of etiologies that can lead to similar abnormal phenotypes. Protein biomarkers are closer to the downstream manifestation of genetic alterations and immune-dysregulation but suffer from manual and subjective interpretations to establish score cut-offs. For example, PD-L1 expression level (i.e., CPS or TPS) offers an imperfect ability to predict treatment response. A significant percentage of potential responders are not identified using existing PD-L1 cut-offs and a portion of PD-L1 positive patients also fail to respond to therapy (1).
The complete picture of disease biology cannot be observed from surveying a very limited amount of specific genetic or protein abnormalities. To unlock new biomarker insights and develop new lifesaving treatments, pharmaceutical manufacturers have been turning to research of the tumor microenvironment (TME) to further evolve an understanding of disease biology.
What is the Tumor Microenvironment?
The TME describes the ecosystem that surrounds and comprises a tumor. (2) Numerous cell and tissue types including immune cells, stromal cells, the blood and vascular networks, as well as various environmental components (e.g., secretory factors), contribute to the regulation of tumor progression and therapeutic response in unique ways. This ecosystem may be differentially represented across disease types and patient populations. There have been various approved therapies directed toward the vasculature, immune checkpoints, and T cells (e.g., aVEGF, PD-L1, CD19) with many treatment targets emerging within the TME (e.g., tertiary lymphoid structures (TLS) and tumor infiltrating lymphocytes (TILs)). (3)
The study of the TME is recognized as one of the most promising areas in oncology drug development to develop a more complete picture of disease biology and tumor progression, particularly around incorporating heterogeneity, morphological patterns, and spatial context into treatment decisions. However, despite the critical role of the TME in tumor progression and regulating treatment efficacy, manual pathology methods have limited the degree to which the TME can be quantified for drug development. Reproducible, scalable, and standardized measurement has been challenging to date due to the sheer amount of data points and time required to systematically characterize each individual cell and feature within the TME.
Utilizing AI Technology to Unlock the TME
AI models have been developed to address many of the problems that have limited research of the TME. Models analyze every pixel of an H&E-stained whole slide image (WSI) to exhaustively quantify and characterize the cellular and tissue composition of the TME. These tumor-specific models are developed and trained utilizing millions of pathologists’ annotations on a set of images. This ensures the accuracy of cell and feature classification and enables a robust quantification of the most salient features for each cancer type.
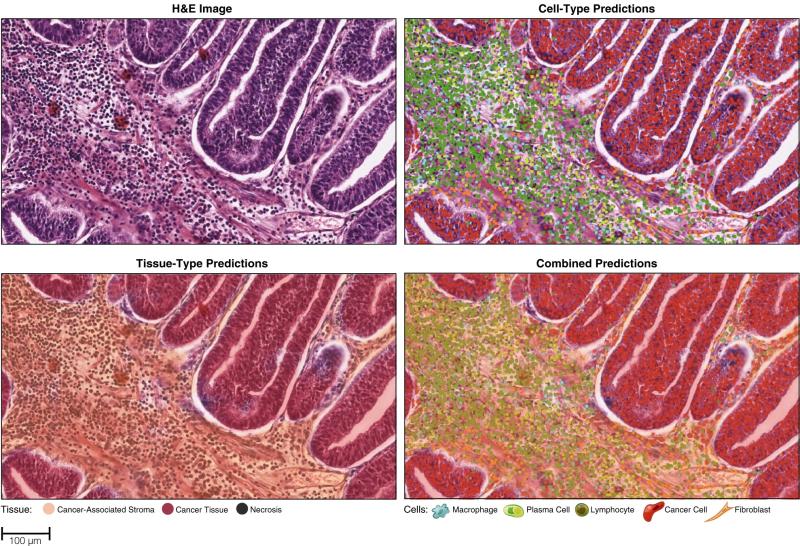
An H&E-stained WSI which an AI model has analyzed and overlayed a colored heatmap depicting visualized predictions of cell-type and tissue-type segmentations. Predictions were then used to compute a diverse array of HIFs for each WSI. (4)
Perhaps the biggest challenge to date has been the ability to utilize and quantify pathology outputs to analyze the TME. Particularly, without a standardized approach to measure the spatial organization of cells and tissue in the TME, analyzing changes within the TME is nearly impossible. As first published in Nature Communications by a team from Harvard, Brigham & Womens’ Hospital, Brown, and PathAI, a standardized approach utilizing human-interpretable features (HIFs) can be used to predict important clinically-relevant molecular phenotypes (HRD score and expression of PD-1, PD-L1, CTLA-4, and TIGIT). HIFs allow researchers to describe counts, areas, densities, and spatial relationships of cell types and tissue compartments across the TME, enabling quantifiable analysis of data at a speed and scale which was previously inaccessible.
Recent Applications of AI in TME Research
AI technology has demonstrated the ability to quantify the TME at different magnifications of analysis ranging from characterizing the TME across several cancer types at once down to assessing each individual cell’s nuclear morphology.
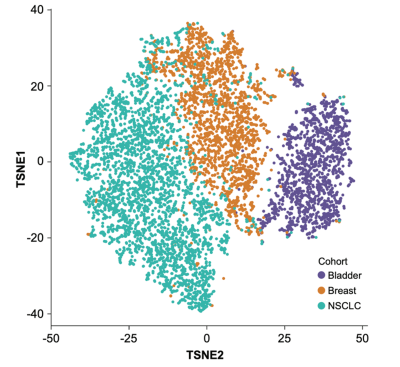
A T-SNE plot showed distinct clusters when an AI model was utilized to stratify H&E samples by cancer type. (5)
AI models can also focus on specific features of interest within the TME, such as molecular signatures. In work presented at AACR 2022, AI models were shown to successfully predict TLS gene expression signatures which correlated with overall survival in breast cancer. (6) At WCLC 2022, results showed AI models can predict c-MET overexpression status in NSCLC with high sensitivity. AI models identified that the density of lymphocytes in cancer epithelium was significantly associated with c-MET positivity. (7) Both use-cases demonstrate that H&E data may have a potential for use as a screening tool to more accurately and efficiently identify patients with molecular signatures which can be targeted for treatment.
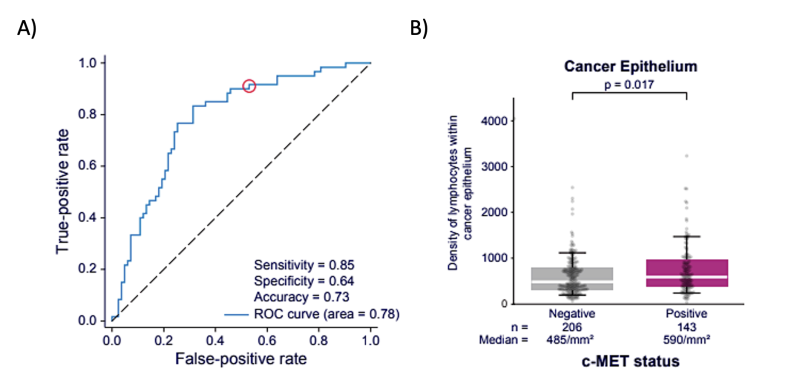
A) AI models predict c-MET over-expression from H&E images with high sensitivity. B) The density of lymphocytes in cancer epithelium is significantly associated with c-MET positivity.(7)
Finally, it has been shown that AI technology is successful at quantifying and analyzing nuclear morphology for predicting patient prognosis. AI models characterizing the TME and nuclear morphology in high-grade serous ovarian cancer identified features of nuclear shape and morphology which were associated with patient prognosis, specifically reduced overall survival. (8)
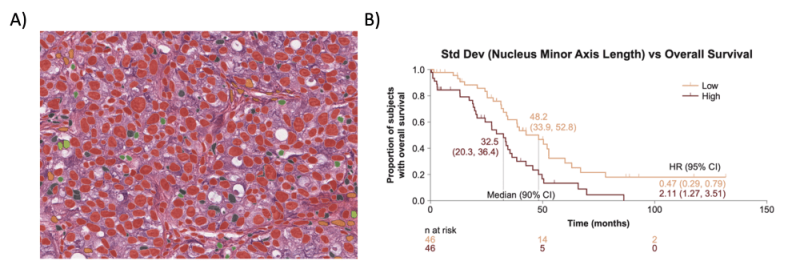
A) An H&E WSI depicting nuclear segmentation across distinct TME cell types. B) Kaplan-Meier survival analysis showing correlation of a feature characterizing the shape of cancer cell nuclei with reduced overall survival in high-grade serous ovarian cancer. (8)
Powering the Next-Generation of Drug Development
Digital pathology is ready to empower the life sciences industry to utilize the TME to develop the next generation of cancer treatments. The countless archives of existing H&E slide images across hospitals and pharmaceuticals could be utilized with TME AI models immediately to potentially discover new biomarkers or the next therapeutic breakthrough. AI technology will continue to improve upon its already impressive accuracy and speed, paving the way for it to eventually becoming the standard go-to diagnostic and pathology tool for drug developers to utilize in their quest to cure cancer.
About PathAI
PathAI is a leading provider of AI-powered research tools and services for pathology. PathAI’s platform promises substantial improvements to the accuracy of diagnosis and the efficacy of treatment of diseases like cancer, leveraging modern approaches in machine and deep learning. Based in Boston, PathAI works with leading life sciences companies and researchers to advance precision medicine. To learn more, visit pathai.com.
To learn more about applications of AI technology in TME research, join PathAI for a FierceBiotech webinar featuring a panel discussion with pharmaceutical leaders.
References
1. Stenehjem DD, Tran D, Nkrumah MA, Gupta S. PD1/PDL1 inhibitors for the treatment of advanced urothelial bladder cancer. Onco Targets Ther. 2018;11:5973-5989 https://doi.org/10.2147/OTT.S135157
2. MD Anderson Cancer Center
3. Leire Bejarano, Marta J.C. Jordāo, Johanna A. Joyce; Therapeutic Targeting of the Tumor Microenvironment. Cancer Discov 1 April 2021; 11 (4): 933–959. https://doi.org/10.1158/2159-8290.CD-20-1808
4. Diao, J.A., Wang, J.K., Chui, W.F. et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat Commun 12, 1613 (2021). https://doi.org/10.1038/s41467-021-21896-9
5. Conway, et al., Generation of an atlas characterizing the tumor immune microenvironment via AI-based histologic mapping of multiple cancer types at scale, Poster presented at Pathology Visions 2022, October 16-18, 2022.
6. Shen, et al., Application of an interpretable graph neural network to predict gene expression signatures associated with tertiary lymphoid structures in histopathological images, Poster presented at the AACR Annual Meeting, April 8-13, 2022. (Abstract 1922)
7. Rajan, et al., Deep-Learning-Based Prediction of c-MET Status From Digitized H&E-Stained Non-Small Cell Lung Cancer Tissue Samples, Poster presented at the IASLC World Conference on Lung Cancer, August 6-9, 2022. (Abstract P2.07-01)
8. Michener, et al., AI-powered analysis of nuclear morphology associated with prognosis in high-grade serous carcinoma, Poster presented at ESMO Congress 2022, September 9-13, 2022. (Abstract 5252)